large_deformation/hyperelastic_tl_up_interactive.py¶
Description
Incompressible Mooney-Rivlin hyperelastic material model.
In this model, the deformation energy density per unit reference volume is given by
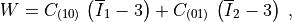
where 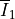 and
and 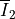 are the first
and second main invariants of the deviatoric part of the right
Cauchy-Green deformation tensor
are the first
and second main invariants of the deviatoric part of the right
Cauchy-Green deformation tensor 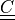 . The coefficients
. The coefficients
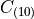 and
and 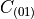 are material parameters.
are material parameters.
Components of the second Piola-Kirchhoff stress are in the case of an incompressible material
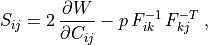
where 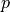 is the hydrostatic pressure.
is the hydrostatic pressure.
The large deformation is described using the total Lagrangian formulation in
this example. The incompressibility is treated by mixed displacement-pressure
formulation. The weak formulation is:
Find the displacement field 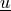 and pressure field
and pressure field 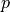 such that:
such that:
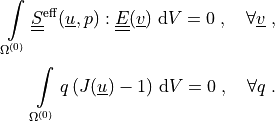
The following formula holds for the axial true (Cauchy) stress in the case of uniaxial stress:
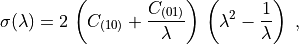
where 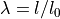 is the prescribed stretch (
is the prescribed stretch (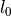 and
and
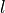 being the original and deformed specimen length respectively).
being the original and deformed specimen length respectively).
The boundary conditions are set so that a state of uniaxial stress is achieved,
i.e. appropriate components of displacement are fixed on the “Left”, “Bottom”,
and “Near” faces and a monotonously increasing displacement is prescribed on
the “Right” face. This prescribed displacement is then used to calculate
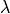 and to convert the second Piola-Kirchhoff stress to the true
(Cauchy) stress.
and to convert the second Piola-Kirchhoff stress to the true
(Cauchy) stress.
Note on material parameters¶
The relationship between material parameters used in the SfePy hyperelastic
terms (NeoHookeanTLTerm,
MooneyRivlinTLTerm)
and the ones used in this example is:
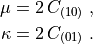
Usage Examples¶
Default options:
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py
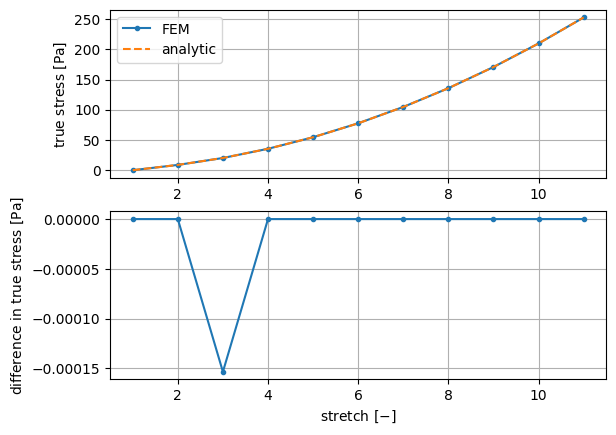
To show a comparison of stress against the analytic formula:
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py -p
Using different mesh fineness:
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
--shape "5, 5, 5"
Different dimensions of the computational domain:
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
--dims "2, 1, 3"
Different length of time interval and/or number of time steps:
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
-t 0,15,21
Use higher approximation order (the -t option to decrease the time step is
required for convergence here):
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
--order 2 -t 0,2,21
Change material parameters:
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py -m 2,1
#!/usr/bin/env python
r"""
Incompressible Mooney-Rivlin hyperelastic material model.
In this model, the deformation energy density per unit reference volume is
given by
.. math::
W = C_{(10)} \, \left( \overline I_1 - 3 \right)
+ C_{(01)} \, \left( \overline I_2 - 3 \right) \;,
where :math:`\overline I_1` and :math:`\overline I_2` are the first
and second main invariants of the deviatoric part of the right
Cauchy-Green deformation tensor :math:`\ull{C}`. The coefficients
:math:`C_{(10)}` and :math:`C_{(01)}` are material parameters.
Components of the second Piola-Kirchhoff stress are in the case of an
incompressible material
.. math::
S_{ij} = 2 \, \pdiff{W}{C_{ij}} - p \, F^{-1}_{ik} \, F^{-T}_{kj} \;,
where :math:`p` is the hydrostatic pressure.
The large deformation is described using the total Lagrangian formulation in
this example. The incompressibility is treated by mixed displacement-pressure
formulation. The weak formulation is:
Find the displacement field :math:`\ul{u}` and pressure field :math:`p`
such that:
.. math::
\intl{\Omega\suz}{} \ull{S}\eff(\ul{u}, p) : \ull{E}(\ul{v})
\difd{V} = 0
\;, \quad \forall \ul{v} \;,
\intl{\Omega\suz}{} q\, (J(\ul{u})-1) \difd{V} = 0
\;, \quad \forall q \;.
The following formula holds for the axial true (Cauchy) stress in the case of
uniaxial stress:
.. math::
\sigma(\lambda) =
2\, \left( C_{(10)} + \frac{C_{(01)}}{\lambda} \right) \,
\left( \lambda^2 - \frac{1}{\lambda} \right) \;,
where :math:`\lambda = l/l_0` is the prescribed stretch (:math:`l_0` and
:math:`l` being the original and deformed specimen length respectively).
The boundary conditions are set so that a state of uniaxial stress is achieved,
i.e. appropriate components of displacement are fixed on the "Left", "Bottom",
and "Near" faces and a monotonously increasing displacement is prescribed on
the "Right" face. This prescribed displacement is then used to calculate
:math:`\lambda` and to convert the second Piola-Kirchhoff stress to the true
(Cauchy) stress.
Note on material parameters
---------------------------
The relationship between material parameters used in the *SfePy* hyperelastic
terms (:class:`NeoHookeanTLTerm
<sfepy.terms.terms_hyperelastic_tl.NeoHookeanTLTerm>`,
:class:`MooneyRivlinTLTerm
<sfepy.terms.terms_hyperelastic_tl.MooneyRivlinTLTerm>`)
and the ones used in this example is:
.. math::
\mu = 2\, C_{(10)} \;,
\kappa = 2\, C_{(01)} \;.
Usage Examples
--------------
Default options::
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py
To show a comparison of stress against the analytic formula::
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py -p
Using different mesh fineness::
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
--shape "5, 5, 5"
Different dimensions of the computational domain::
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
--dims "2, 1, 3"
Different length of time interval and/or number of time steps::
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
-t 0,15,21
Use higher approximation order (the ``-t`` option to decrease the time step is
required for convergence here)::
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py \
--order 2 -t 0,2,21
Change material parameters::
$ python sfepy/examples/large_deformation/hyperelastic_tl_up_interactive.py -m 2,1
"""
import argparse
import sys
SFEPY_DIR = '.'
sys.path.append(SFEPY_DIR)
import matplotlib.pyplot as plt
import numpy as np
from sfepy.base.base import IndexedStruct, Struct
from sfepy.discrete import (
FieldVariable, Material, Integral, Function, Equation, Equations, Problem)
from sfepy.discrete.conditions import Conditions, EssentialBC
from sfepy.discrete.fem import FEDomain, Field
from sfepy.homogenization.utils import define_box_regions
from sfepy.mesh.mesh_generators import gen_block_mesh
from sfepy.solvers.ls import ScipyDirect
from sfepy.solvers.nls import Newton
from sfepy.solvers.ts_solvers import SimpleTimeSteppingSolver
from sfepy.terms import Term
DIMENSION = 3
def get_displacement(ts, coors, bc=None, problem=None):
"""
Define the time-dependent displacement.
"""
out = 1. * ts.time * coors[:, 0]
return out
def plot_graphs(
material_parameters, global_stress, global_displacement,
undeformed_length):
"""
Plot a comparison of the true stress computed by the FEM and using the
analytic formula.
Parameters
----------
material_parameters : list or tuple of float
The C10 and C01 coefficients.
global_displacement
The total displacement for each time step, from the FEM.
global_stress
The true (Cauchy) stress for each time step, from the FEM.
undeformed_length : float
The length of the undeformed specimen.
"""
c10, c01 = material_parameters
stretch = 1 + np.array(global_displacement) / undeformed_length
# axial stress values
stress_fem_2pk = np.array([sig for sig in global_stress])
stress_fem = stress_fem_2pk * stretch**2
stress_analytic = 2 * (c10 + c01/stretch) * (stretch**2 - 1./stretch)
fig = plt.figure()
ax_stress = fig.add_subplot(211)
ax_difference = fig.add_subplot(212)
ax_stress.plot(stretch, stress_fem, '.-', label='FEM')
ax_stress.plot(stretch, stress_analytic, '--', label='analytic')
ax_difference.plot(stretch, stress_fem - stress_analytic, '.-')
ax_stress.legend(loc='best').set_draggable(True)
ax_stress.set_ylabel(r'true stress $\mathrm{[Pa]}$')
ax_stress.grid()
ax_difference.set_ylabel(r'difference in true stress $\mathrm{[Pa]}$')
ax_difference.set_xlabel(r'stretch $\mathrm{[-]}$')
ax_difference.grid()
return fig
def stress_strain(
out, problem, _state, order=1, global_stress=None,
global_displacement=None, **_):
"""
Compute the stress and the strain and add them to the output.
Parameters
----------
out : dict
Holds the results of the finite element computation.
problem : sfepy.discrete.Problem
order : int
The approximation order of the displacement field.
global_displacement
Total displacement for each time step, current value will be appended.
global_stress
The true (Cauchy) stress for each time step, current value will be
appended.
Returns
-------
out : dict
"""
strain = problem.evaluate(
'dw_tl_he_neohook.%d.Omega(m.mu, v, u)' % (2*order),
mode='el_avg', term_mode='strain', copy_materials=False)
out['green_strain'] = Struct(
name='output_data', mode='cell', data=strain, dofs=None)
stress_10 = problem.evaluate(
'dw_tl_he_neohook.%d.Omega(m.mu, v, u)' % (2*order),
mode='el_avg', term_mode='stress', copy_materials=False)
stress_01 = problem.evaluate(
'dw_tl_he_mooney_rivlin.%d.Omega(m.kappa, v, u)' % (2*order),
mode='el_avg', term_mode='stress', copy_materials=False)
stress_p = problem.evaluate(
'dw_tl_bulk_pressure.%d.Omega(v, u, p)' % (2*order),
mode='el_avg', term_mode='stress', copy_materials=False)
stress = stress_10 + stress_01 + stress_p
out['stress'] = Struct(
name='output_data', mode='cell', data=stress, dofs=None)
global_stress.append(stress[0, 0, 0, 0])
global_displacement.append(np.max(out['u'].data[:, 0]))
return out
def main(cli_args):
dims = parse_argument_list(cli_args.dims, float)
shape = parse_argument_list(cli_args.shape, int)
centre = parse_argument_list(cli_args.centre, float)
material_parameters = parse_argument_list(cli_args.material_parameters,
float)
order = cli_args.order
ts_vals = cli_args.ts.split(',')
ts = {
't0' : float(ts_vals[0]), 't1' : float(ts_vals[1]),
'n_step' : int(ts_vals[2])}
do_plot = cli_args.plot
### Mesh and regions ###
mesh = gen_block_mesh(
dims, shape, centre, name='block', verbose=False)
domain = FEDomain('domain', mesh)
omega = domain.create_region('Omega', 'all')
lbn, rtf = domain.get_mesh_bounding_box()
box_regions = define_box_regions(3, lbn, rtf)
regions = dict([
[r, domain.create_region(r, box_regions[r][0], box_regions[r][1])]
for r in box_regions])
### Fields ###
scalar_field = Field.from_args(
'fu', np.float64, 'scalar', omega, approx_order=order-1)
vector_field = Field.from_args(
'fv', np.float64, 'vector', omega, approx_order=order)
u = FieldVariable('u', 'unknown', vector_field, history=1)
v = FieldVariable('v', 'test', vector_field, primary_var_name='u')
p = FieldVariable('p', 'unknown', scalar_field, history=1)
q = FieldVariable('q', 'test', scalar_field, primary_var_name='p')
### Material ###
c10, c01 = material_parameters
m = Material(
'm', mu=2*c10, kappa=2*c01,
)
### Boundary conditions ###
x_sym = EssentialBC('x_sym', regions['Left'], {'u.0' : 0.0})
y_sym = EssentialBC('y_sym', regions['Near'], {'u.1' : 0.0})
z_sym = EssentialBC('z_sym', regions['Bottom'], {'u.2' : 0.0})
disp_fun = Function('disp_fun', get_displacement)
displacement = EssentialBC(
'displacement', regions['Right'], {'u.0' : disp_fun})
ebcs = Conditions([x_sym, y_sym, z_sym, displacement])
### Terms and equations ###
integral = Integral('i', order=2*order)
term_neohook = Term.new(
'dw_tl_he_neohook(m.mu, v, u)',
integral, omega, m=m, v=v, u=u)
term_mooney = Term.new(
'dw_tl_he_mooney_rivlin(m.kappa, v, u)',
integral, omega, m=m, v=v, u=u)
term_pressure = Term.new(
'dw_tl_bulk_pressure(v, u, p)',
integral, omega, v=v, u=u, p=p)
term_volume_change = Term.new(
'dw_tl_volume(q, u)',
integral, omega, q=q, u=u, term_mode='volume')
term_volume = Term.new(
'dw_integrate(q)',
integral, omega, q=q)
eq_balance = Equation('balance', term_neohook+term_mooney+term_pressure)
eq_volume = Equation('volume', term_volume_change-term_volume)
equations = Equations([eq_balance, eq_volume])
### Solvers ###
ls = ScipyDirect({})
nls_status = IndexedStruct()
nls = Newton(
{'i_max' : 5},
lin_solver=ls, status=nls_status
)
### Problem ###
pb = Problem('hyper', equations=equations)
pb.set_bcs(ebcs=ebcs)
pb.set_ics(ics=Conditions([]))
tss = SimpleTimeSteppingSolver(ts, nls=nls, context=pb)
pb.set_solver(tss)
### Solution ###
axial_stress = []
axial_displacement = []
def stress_strain_fun(*args, **kwargs):
return stress_strain(
*args, order=order, global_stress=axial_stress,
global_displacement=axial_displacement, **kwargs)
pb.solve(save_results=True, post_process_hook=stress_strain_fun)
if do_plot:
fig = plot_graphs(
material_parameters, axial_stress, axial_displacement,
undeformed_length=dims[0])
fig.savefig('hyperelastic_tl_up_comparison.png', bbox_inches='tight')
if cli_args.show:
plt.show()
def parse_argument_list(cli_arg, type_fun=None, value_separator=','):
"""
Split the command-line argument into a list of items of given type.
Parameters
----------
cli_arg : str
type_fun : function
A function to be called on each substring of `cli_arg`; default: str.
value_separator : str
"""
if type_fun is None:
type_fun = str
out = [type_fun(value) for value in cli_arg.split(value_separator)]
return out
def parse_args():
"""Parse command line arguments."""
parser = argparse.ArgumentParser(
description=__doc__,
formatter_class=argparse.RawDescriptionHelpFormatter)
parser.add_argument(
'--order', type=int, default=1, help='The approximation order of the '
'displacement field [default: %(default)s]')
parser.add_argument(
'-m', '--material-parameters', default='1.0, 0.5',
help='Material parameters - C10, C01 - of the two-parametric '
'Mooney-Rivlin hyperelastic model. [default: %(default)s]')
parser.add_argument(
'--dims', default="1.0, 1.0, 1.0",
help='Dimensions of the block [default: %(default)s]')
parser.add_argument(
'--shape', default='4, 4, 4',
help='Shape (counts of nodes in x, y, z) of the block [default: '
'%(default)s]')
parser.add_argument(
'--centre', default='0.5, 0.5, 0.5',
help='Centre of the block [default: %(default)s]')
parser.add_argument(
'-p', '--plot', action='store_true', default=False,
help='Whether to plot a comparison with analytical formula.')
parser.add_argument(
'-n', '--no-show', dest='show', action='store_false', default=True,
help='Do not show matplotlib figures.')
parser.add_argument(
'-t', '--ts',
type=str, default='0.0,10.0,11',
help='Start time, end time, and number of time steps [default: '
'"%(default)s"]')
return parser.parse_args()
if __name__ == '__main__':
args = parse_args()
main(args)