large_deformation/hyperelastic_ul.py¶
Description
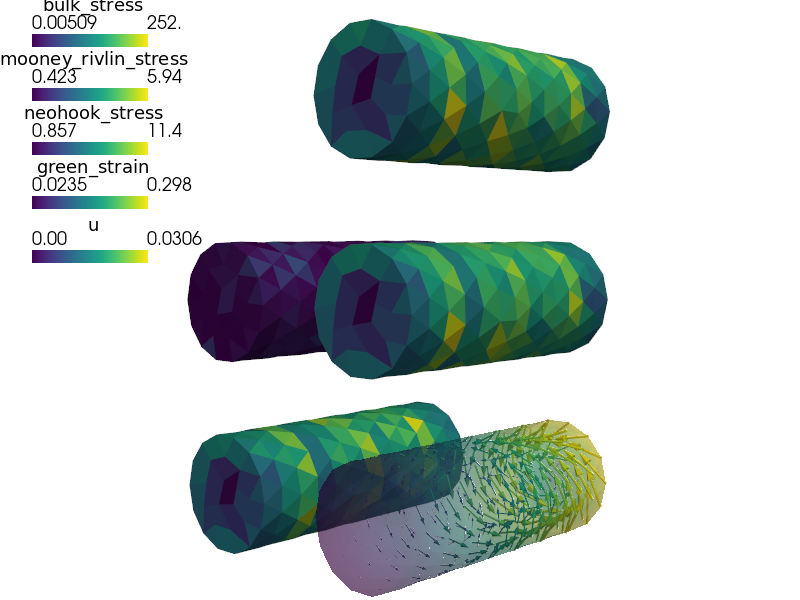
Nearly incompressible Mooney-Rivlin hyperelastic material model.
Large deformation is described using the updated Lagrangian formulation. Models of this kind can be used to model e.g. rubber or some biological materials.
# -*- coding: utf-8 -*-
r"""
Nearly incompressible Mooney-Rivlin hyperelastic material model.
Large deformation is described using the updated Lagrangian formulation.
Models of this kind can be used to model e.g. rubber or some biological
materials.
"""
import numpy as nm
from sfepy import data_dir
filename_mesh = data_dir + '/meshes/3d/cylinder.mesh'
options = {
'nls': 'newton',
'ls': 'ls',
'ts': 'ts',
'ulf': True,
'mesh_update_variables': ['u'],
'output_dir': 'output',
'post_process_hook': 'stress_strain',
'report_nls_status': True,
'log_nls_status': True,
}
fields = {
'displacement': ('real', 3, 'Omega', 1),
}
materials = {
'solid': ({'K': 1e3, # bulk modulus
'mu': 20e0, # shear modulus of neoHookean term
'kappa': 10e0, # shear modulus of Mooney-Rivlin term
},),
}
variables = {
'u': ('unknown field', 'displacement', 0),
'v': ('test field', 'displacement', 'u'),
}
regions = {
'Omega' : 'all',
'Left' : ('vertices in (x < 0.001)', 'facet'),
'Right' : ('vertices in (x > 0.099)', 'facet'),
}
##
# Dirichlet BC + related functions.
ebcs = {
'l' : ('Left', {'u.all' : 0.0}),
'r' : ('Right', {'u.0' : 0.0, 'u.[1,2]' : 'rotate_yz'}),
}
centre = nm.array( [0, 0], dtype = nm.float64 )
def rotate_yz(ts, coor, **kwargs):
from sfepy.linalg import rotation_matrix2d
vec = coor[:,1:3] - centre
angle = 10.0 * ts.step
print('angle:', angle)
mtx = rotation_matrix2d( angle )
vec_rotated = nm.dot( vec, mtx )
displacement = vec_rotated - vec
return displacement
functions = {
'rotate_yz' : (rotate_yz,),
}
def stress_strain( out, problem, state, extend = False ):
from sfepy.base.base import Struct
ev = problem.evaluate
strain = ev('dw_ul_he_neohook.3.Omega( solid.mu, v, u )',
mode='el_avg', term_mode='strain')
out['green_strain'] = Struct(name='output_data',
mode='cell', data=strain, dofs=None)
stress = ev('dw_ul_he_neohook.3.Omega( solid.mu, v, u )',
mode='el_avg', term_mode='stress')
out['neohook_stress'] = Struct(name='output_data',
mode='cell', data=stress, dofs=None)
stress = ev('dw_ul_he_mooney_rivlin.3.Omega( solid.kappa, v, u )',
mode='el_avg', term_mode='stress')
out['mooney_rivlin_stress'] = Struct(name='output_data',
mode='cell', data=stress, dofs=None)
stress = ev('dw_ul_bulk_penalty.3.Omega( solid.K, v, u )',
mode='el_avg', term_mode= 'stress')
out['bulk_stress'] = Struct(name='output_data',
mode='cell', data=stress, dofs=None)
return out
equations = {
'balance': """dw_ul_he_neohook.3.Omega( solid.mu, v, u )
+ dw_ul_he_mooney_rivlin.3.Omega(solid.kappa, v, u)
+ dw_ul_bulk_penalty.3.Omega( solid.K, v, u )
= 0""",
}
##
# Solvers etc.
solvers = {
'ls': ('ls.scipy_direct', {}),
'newton': ('nls.newton', {
'i_max': 25,
'eps_a': 1e-8,
'eps_r': 1.0,
'macheps': 1e-16,
'lin_red': 1e-2, # Linear system error < (eps_a * lin_red).
'ls_red': 0.1,
'ls_red_warp': 0.001,
'ls_on': 1.1,
'ls_min': 1e-5,
'check': 0,
'delta': 1e-6,
}),
'ts': ('ts.simple', {
't0': 0,
't1': 1,
'dt': None,
'n_step': 11, # has precedence over dt!
'verbose' : 1,
}),
}