diffusion/laplace_iga_interactive.py¶
Description
Laplace equation with Dirichlet boundary conditions solved in a single patch NURBS domain using the isogeometric analysis (IGA) approach, using commands for interactive use.
This script allows the creation of a customisable NURBS surface using igakit built-in CAD routines, which is then saved in custom HDF5-based files with .iga extension.
Notes¶
The create_patch function creates a NURBS-patch of the area between two
coplanar nested circles using igakit CAD built-in routines. The created patch
is not connected in the orthoradial direction. This is a problem when the
disconnected boundary is not perpendicular to the line connecting the two
centres of the circles, as the solution then exhibits a discontinuity along
this line. A workaround for this issue is to enforce perpendicularity by
changing the start angle in function igakit.cad.circle (see the code down
below for the actual trick). The discontinuity disappears.
Usage Examples¶
Default options, storing results in this file’s parent directory:
python3 sfepy/examples/diffusion/laplace_iga_interactive.py
Command line options for tweaking the geometry of the NURBS-patch & more:
python3 sfepy/examples/diffusion/laplace_iga_interactive.py --R1=0.7 --C2=0.1,0.1 --viewpatch
View the results using:
sfepy-view concentric_circles.vtk
#!/usr/bin/env python
r"""
Laplace equation with Dirichlet boundary conditions solved in a single patch
NURBS domain using the isogeometric analysis (IGA) approach, using commands
for interactive use.
This script allows the creation of a customisable NURBS surface using igakit
built-in CAD routines, which is then saved in custom HDF5-based files with
.iga extension.
Notes
-----
The ``create_patch`` function creates a NURBS-patch of the area between two
coplanar nested circles using igakit CAD built-in routines. The created patch
is not connected in the orthoradial direction. This is a problem when the
disconnected boundary is not perpendicular to the line connecting the two
centres of the circles, as the solution then exhibits a discontinuity along
this line. A workaround for this issue is to enforce perpendicularity by
changing the start angle in function ``igakit.cad.circle`` (see the code down
below for the actual trick). The discontinuity disappears.
Usage Examples
--------------
Default options, storing results in this file's parent directory::
python3 sfepy/examples/diffusion/laplace_iga_interactive.py
Command line options for tweaking the geometry of the NURBS-patch & more::
python3 sfepy/examples/diffusion/laplace_iga_interactive.py --R1=0.7 --C2=0.1,0.1 --viewpatch
View the results using::
sfepy-view concentric_circles.vtk
"""
from argparse import RawDescriptionHelpFormatter, ArgumentParser
import os
import sys
sys.path.append('.')
import numpy as nm
from sfepy import data_dir
from sfepy.base.ioutils import ensure_path
from sfepy.base.base import IndexedStruct
from sfepy.discrete import (FieldVariable, Integral, Equation,Equations,
Problem)
from sfepy.discrete.iga.domain import IGDomain
from sfepy.discrete.common.fields import Field
from sfepy.terms import Term
from sfepy.discrete.conditions import Conditions, EssentialBC
from sfepy.solvers.ls import ScipyDirect
from sfepy.solvers.nls import Newton
def create_patch(R1, R2, C1, C2, order=2, viewpatch=False):
"""
Create a single 2d NURBS-patch of the area between two coplanar nested
circles using igakit.
Parameters
----------
R1 : float
Radius of the inner circle.
R2 : float
Radius of the outer circle.
C1 : list of two floats
Coordinates of the center of the inner circle given as [x1, y1].
C2 : list of two floats
Coordinates of the center of the outer circle given as [x2, y2].
order : int, optional
Degree of the NURBS basis functions. The default is 2.
viewpatch : bool, optional
When set to True, display the NURBS patch. The default is False.
Returns
-------
None.
"""
from sfepy.discrete.iga.domain_generators import create_from_igakit
import sfepy.discrete.iga.io as io
from igakit.cad import circle, ruled
from igakit.plot import plt as iplt
from numpy import pi
# Assert the inner circle is inside the outer one
inter_centers = nm.sqrt((C2[0]-C1[0])**2 + (C2[1]-C1[1])**2)
assert R2>R1, "Outer circle should have a larger radius than the inner one"
assert inter_centers<R2-R1, "Circles are not nested"
# Geometry Creation
centers_direction = [C2[0]-C1[0], C2[1]-C1[1]]
if centers_direction[0]==0 and centers_direction[1]==0:
start_angle = 0.0
else:
start_angle = nm.arctan2(centers_direction[1], centers_direction[0])
c1 = circle(radius=R1, center=C1, angle=(start_angle, start_angle + 2*pi))
c2 = circle(radius=R2, center=C2, angle=(start_angle, start_angle + 2*pi))
srf = ruled(c1,c2).transpose() # make the radial direction first
# Refinement
insert_U = insert_uniformly(srf.knots[0], 6)
insert_V = insert_uniformly(srf.knots[1], 6)
srf.refine(0, insert_U).refine(1, insert_V)
# Setting the NURBS-surface degree
srf.elevate(0, order-srf.degree[0] if order-srf.degree[0] > 0 else 0)
srf.elevate(1, order-srf.degree[1] if order-srf.degree[1] > 0 else 0)
# Sfepy .iga file creation
nurbs, bmesh, regions = create_from_igakit(srf, verbose=True)
# Save .iga file in sfepy/meshes/iga
filename_domain = data_dir + '/meshes/iga/concentric_circles.iga'
io.write_iga_data(filename_domain, None, nurbs.knots, nurbs.degrees,
nurbs.cps, nurbs.weights, nurbs.cs, nurbs.conn,
bmesh.cps, bmesh.weights, bmesh.conn, regions)
if viewpatch:
iplt.use('matplotlib')
iplt.figure()
iplt.plot(srf)
iplt.show()
def insert_uniformly(U, n):
"""
Find knots to uniformly add to U.
[Code from igakit/demo/venturi.py file]
Given a knot vector U and the number of uniform spans desired,
find the knots which need to be inserted.
Parameters
----------
U : numpy.ndarray
Original knot vector for a C^p-1 space.
n : int
Target number of uniformly-spaced knot spans.
Returns
-------
Knots to be inserted into U
"""
U0 = U
dU=(U.max()-U.min())/float(n) # target dU in knot vector
idone=0
while idone == 0:
# Add knots in middle of spans which are too large
Uadd=[]
for i in range(len(U)-1):
if U[i+1]-U[i] > dU:
Uadd.append(0.5*(U[i+1]+U[i]))
# Now we add these knots (once only, assumes C^(p-1))
if len(Uadd) > 0:
U = nm.sort(nm.concatenate([U,nm.asarray(Uadd)]))
else:
idone=1
# And now a little Laplacian smoothing
for num_iterations in range(5):
for i in range(len(U)-2):
if abs(U0[U0.searchsorted(U[i+1])]-U[i+1]) > 1.0e-14:
U[i+1] = 0.5*(U[i]+U[i+2])
return nm.setdiff1d(U,U0)
helps = {
'output_dir' :
'output directory',
'R1' :
'Inner circle radius [default: %(default)s]',
'R2' :
'Outer circle radius [default: %(default)s]',
'C1' :
'centre of the inner circle [default: %(default)s]',
'C2' :
'centre of the outer circle [default: %(default)s]',
'order' :
'field approximation order [default: %(default)s]',
'viewpatch' :
'generate a plot of the NURBS-patch',
}
def main():
parser = ArgumentParser(description=__doc__.rstrip(),
formatter_class=RawDescriptionHelpFormatter)
parser.add_argument('-o', '--output-dir', default='.',
help=helps['output_dir'])
parser.add_argument('--R1', metavar='R1',
action='store', dest='R1',
default='0.5', help=helps['R1'])
parser.add_argument('--R2', metavar='R2',
action='store', dest='R2',
default='1.0', help=helps['R2'])
parser.add_argument('--C1', metavar='C1',
action='store', dest='C1',
default='0.0,0.0', help=helps['C1'])
parser.add_argument('--C2', metavar='C2',
action='store', dest='C2',
default='0.0,0.0', help=helps['C2'])
parser.add_argument('--order', metavar='int', type=int,
action='store', dest='order',
default=2, help=helps['order'])
parser.add_argument('-v', '--viewpatch',
action='store_true', dest='viewpatch',
default=False, help=helps['viewpatch'])
options = parser.parse_args()
# Creation of the NURBS-patch with igakit
R1 = eval(options.R1)
R2 = eval(options.R2)
C1 = list(eval(options.C1))
C2 = list(eval(options.C2))
order = options.order
viewpatch = options.viewpatch
create_patch(R1, R2, C1, C2, order=order, viewpatch=viewpatch)
# Setting a Domain instance
filename_domain = data_dir + '/meshes/iga/concentric_circles.iga'
domain = IGDomain.from_file(filename_domain)
# Sub-domains
omega = domain.create_region('Omega', 'all')
Gamma_out = domain.create_region('Gamma_out', 'vertices of set xi01',
kind='facet')
Gamma_in = domain.create_region('Gamma_in', 'vertices of set xi00',
kind='facet')
# Field (featuring order elevation)
order_increase = order - domain.nurbs.degrees[0]
order_increase *= int(order_increase>0)
field = Field.from_args('fu', nm.float64, 'scalar', omega,
approx_order='iga', space='H1',
poly_space_basis='iga')
# Variables
u = FieldVariable('u', 'unknown', field) # unknown function
v = FieldVariable('v', 'test', field, primary_var_name='u') # test function
# Integral
integral = Integral('i', order=2*field.approx_order)
# Term
t = Term.new('dw_laplace( v, u )', integral, omega, v=v, u=u)
# Equation
eq = Equation('laplace', t)
eqs = Equations([eq])
# Boundary Conditions
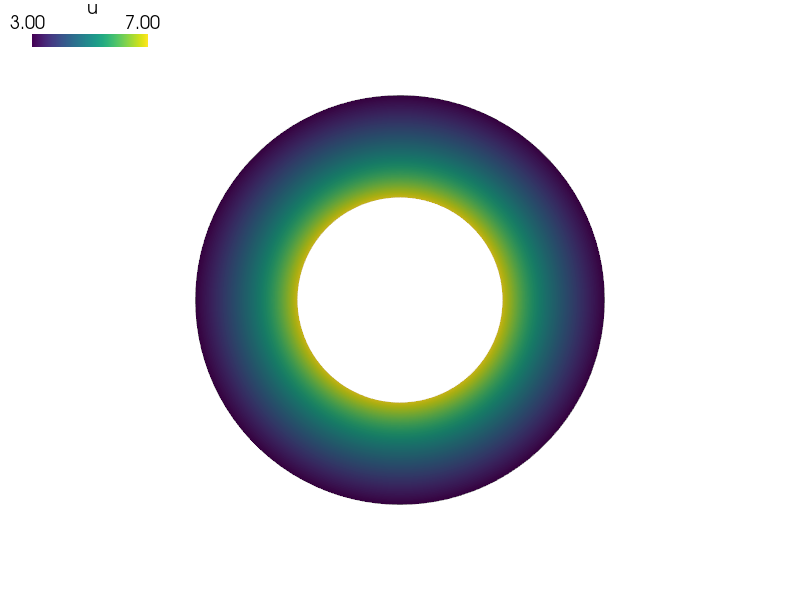
u_in = EssentialBC('u_in', Gamma_in, {'u.all' : 7.0})
u_out = EssentialBC('u_out', Gamma_out, {'u.all' : 3.0})
# solvers
ls = ScipyDirect({})
nls_status = IndexedStruct()
nls = Newton({}, lin_solver=ls, status=nls_status)
# problem instance
pb = Problem('potential', equations=eqs, active_only=True)
# Set boundary conditions
pb.set_bcs(ebcs=Conditions([u_in, u_out]))
# solving
pb.set_solver(nls)
status = IndexedStruct()
state = pb.solve(status=status, save_results=True, verbose=True)
# Saving the results to a classic VTK file
filename = os.path.join(options.output_dir, 'concentric_circles.vtk')
ensure_path(filename)
pb.save_state(filename, state)
if __name__ == '__main__':
main()