multi_physics/biot_parallel_interactive.py¶
Description
Parallel assembling and solving of a Biot problem (deformable porous medium), using commands for interactive use.
Find 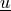 ,
, 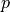 such that:
such that:
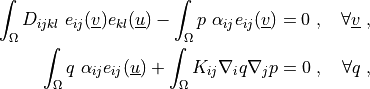
where
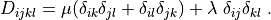
Important Notes¶
This example requires petsc4py, mpi4py and (optionally) pymetis with their dependencies installed!
This example generates a number of files - do not use an existing non-empty directory for the
output_dirargument.Use the
--clearoption with care!
Notes¶
Each task is responsible for a subdomain consisting of a set of cells (a cell region).
Each subdomain owns PETSc DOFs within a consecutive range.
When both global and task-local variables exist, the task-local variables have
_isuffix.This example shows how to use a nonlinear solver from PETSc.
This example can serve as a template for solving a (non)linear multi-field problem - just replace the equations in
create_local_problem().The material parameter
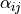 is artificially high to be able to
see the pressure influence on displacements.
is artificially high to be able to
see the pressure influence on displacements.The command line options are saved into <output_dir>/options.txt file.
Usage Examples¶
See all options:
python3 sfepy/examples/multi_physics/biot_parallel_interactive.py -h
See PETSc options:
python3 sfepy/examples/multi_physics/biot_parallel_interactive.py -help
Single process run useful for debugging with debug():
python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel
Parallel runs:
mpiexec -n 3 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel -2 --shape=101,101
mpiexec -n 3 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel -2 --shape=101,101 --metis
mpiexec -n 8 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel -2 --shape 101,101 --metis -snes_monitor -snes_view -snes_converged_reason -ksp_monitor
Using FieldSplit preconditioner:
mpiexec -n 2 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel --shape=101,101 -snes_monitor -snes_converged_reason -ksp_monitor -pc_type fieldsplit
mpiexec -n 8 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel --shape=1001,1001 --metis -snes_monitor -snes_converged_reason -ksp_monitor -pc_type fieldsplit -pc_fieldsplit_type additive
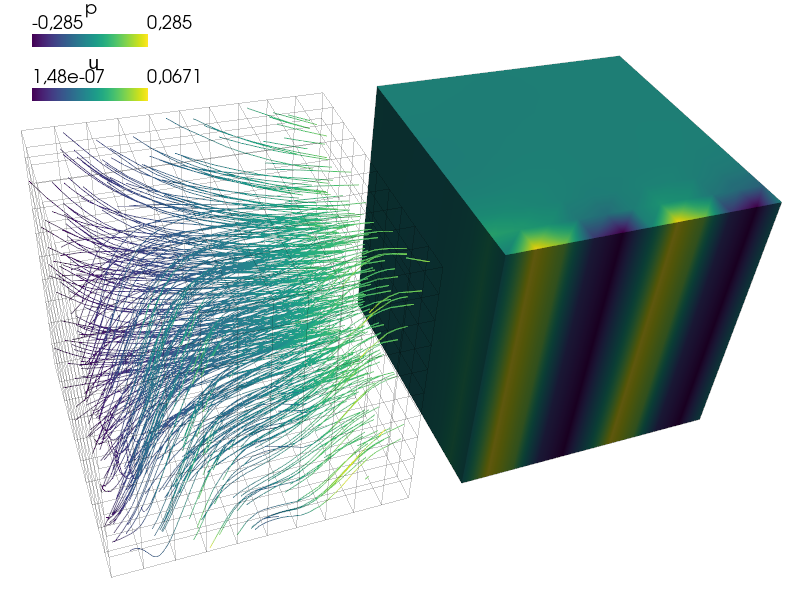
View the results using:
sfepy-view output-parallel/sol.h5
#!/usr/bin/env python
r"""
Parallel assembling and solving of a Biot problem (deformable porous medium),
using commands for interactive use.
Find :math:`\ul{u}`, :math:`p` such that:
.. math::
\int_{\Omega} D_{ijkl}\ e_{ij}(\ul{v}) e_{kl}(\ul{u})
- \int_{\Omega} p\ \alpha_{ij} e_{ij}(\ul{v})
= 0
\;, \quad \forall \ul{v} \;,
\int_{\Omega} q\ \alpha_{ij} e_{ij}(\ul{u})
+ \int_{\Omega} K_{ij} \nabla_i q \nabla_j p
= 0
\;, \quad \forall q \;,
where
.. math::
D_{ijkl} = \mu (\delta_{ik} \delta_{jl}+\delta_{il} \delta_{jk}) +
\lambda \ \delta_{ij} \delta_{kl}
\;.
Important Notes
---------------
- This example requires petsc4py, mpi4py and (optionally) pymetis with their
dependencies installed!
- This example generates a number of files - do not use an existing non-empty
directory for the ``output_dir`` argument.
- Use the ``--clear`` option with care!
Notes
-----
- Each task is responsible for a subdomain consisting of a set of cells (a cell
region).
- Each subdomain owns PETSc DOFs within a consecutive range.
- When both global and task-local variables exist, the task-local
variables have ``_i`` suffix.
- This example shows how to use a nonlinear solver from PETSc.
- This example can serve as a template for solving a (non)linear multi-field
problem - just replace the equations in :func:`create_local_problem()`.
- The material parameter :math:`\alpha_{ij}` is artificially high to be able to
see the pressure influence on displacements.
- The command line options are saved into <output_dir>/options.txt file.
Usage Examples
--------------
See all options::
python3 sfepy/examples/multi_physics/biot_parallel_interactive.py -h
See PETSc options::
python3 sfepy/examples/multi_physics/biot_parallel_interactive.py -help
Single process run useful for debugging with :func:`debug()
<sfepy.base.base.debug>`::
python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel
Parallel runs::
mpiexec -n 3 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel -2 --shape=101,101
mpiexec -n 3 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel -2 --shape=101,101 --metis
mpiexec -n 8 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel -2 --shape 101,101 --metis -snes_monitor -snes_view -snes_converged_reason -ksp_monitor
Using FieldSplit preconditioner::
mpiexec -n 2 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel --shape=101,101 -snes_monitor -snes_converged_reason -ksp_monitor -pc_type fieldsplit
mpiexec -n 8 python3 sfepy/examples/multi_physics/biot_parallel_interactive.py output-parallel --shape=1001,1001 --metis -snes_monitor -snes_converged_reason -ksp_monitor -pc_type fieldsplit -pc_fieldsplit_type additive
View the results using::
sfepy-view output-parallel/sol.h5
"""
from argparse import RawDescriptionHelpFormatter, ArgumentParser
import os
import sys
sys.path.append('.')
import numpy as nm
from sfepy.base.base import output, Struct
from sfepy.base.ioutils import ensure_path, remove_files_patterns, save_options
from sfepy.base.timing import Timer
from sfepy.discrete.fem import Mesh, FEDomain, Field
from sfepy.discrete.common.region import Region
from sfepy.discrete import (FieldVariable, Material, Integral, Function,
Equation, Equations, Problem)
from sfepy.discrete.conditions import Conditions, EssentialBC
from sfepy.terms import Term
from sfepy.solvers.ls import PETScKrylovSolver
from sfepy.solvers.nls import PETScNonlinearSolver
from sfepy.mechanics.matcoefs import stiffness_from_lame
import sfepy.parallel.parallel as pl
from sfepy.parallel.evaluate import PETScParallelEvaluator
def create_local_problem(omega_gi, orders):
"""
Local problem definition using a domain corresponding to the global region
`omega_gi`.
"""
order_u, order_p = orders
mesh = omega_gi.domain.mesh
# All tasks have the whole mesh.
bbox = mesh.get_bounding_box()
min_x, max_x = bbox[:, 0]
eps_x = 1e-8 * (max_x - min_x)
min_y, max_y = bbox[:, 1]
eps_y = 1e-8 * (max_y - min_y)
mesh_i = Mesh.from_region(omega_gi, mesh, localize=True)
domain_i = FEDomain('domain_i', mesh_i)
omega_i = domain_i.create_region('Omega', 'all')
gamma1_i = domain_i.create_region('Gamma1',
'vertices in (x < %.10f)'
% (min_x + eps_x),
'facet', allow_empty=True)
gamma2_i = domain_i.create_region('Gamma2',
'vertices in (x > %.10f)'
% (max_x - eps_x),
'facet', allow_empty=True)
gamma3_i = domain_i.create_region('Gamma3',
'vertices in (y < %.10f)'
% (min_y + eps_y),
'facet', allow_empty=True)
field1_i = Field.from_args('fu', nm.float64, mesh.dim, omega_i,
approx_order=order_u)
field2_i = Field.from_args('fp', nm.float64, 1, omega_i,
approx_order=order_p)
output('field 1: number of local DOFs:', field1_i.n_nod)
output('field 2: number of local DOFs:', field2_i.n_nod)
u_i = FieldVariable('u_i', 'unknown', field1_i, order=0)
v_i = FieldVariable('v_i', 'test', field1_i, primary_var_name='u_i')
p_i = FieldVariable('p_i', 'unknown', field2_i, order=1)
q_i = FieldVariable('q_i', 'test', field2_i, primary_var_name='p_i')
if mesh.dim == 2:
alpha = 1e2 * nm.array([[0.132], [0.132], [0.092]])
else:
alpha = 1e2 * nm.array([[0.132], [0.132], [0.132],
[0.092], [0.092], [0.092]])
mat = Material('m', D=stiffness_from_lame(mesh.dim, lam=10, mu=5),
k=1, alpha=alpha)
integral = Integral('i', order=2*(max(order_u, order_p)))
t11 = Term.new('dw_lin_elastic(m.D, v_i, u_i)',
integral, omega_i, m=mat, v_i=v_i, u_i=u_i)
t12 = Term.new('dw_biot(m.alpha, v_i, p_i)',
integral, omega_i, m=mat, v_i=v_i, p_i=p_i)
t21 = Term.new('dw_biot(m.alpha, u_i, q_i)',
integral, omega_i, m=mat, u_i=u_i, q_i=q_i)
t22 = Term.new('dw_laplace(m.k, q_i, p_i)',
integral, omega_i, m=mat, q_i=q_i, p_i=p_i)
eq1 = Equation('eq1', t11 - t12)
eq2 = Equation('eq1', t21 + t22)
eqs = Equations([eq1, eq2])
ebc1 = EssentialBC('ebc1', gamma1_i, {'u_i.all' : 0.0})
ebc2 = EssentialBC('ebc2', gamma2_i, {'u_i.0' : 0.05})
def bc_fun(ts, coors, **kwargs):
val = 0.3 * nm.sin(4 * nm.pi * (coors[:, 0] - min_x) / (max_x - min_x))
return val
fun = Function('bc_fun', bc_fun)
ebc3 = EssentialBC('ebc3', gamma3_i, {'p_i.all' : fun})
pb = Problem('problem_i', equations=eqs, active_only=False)
pb.time_update(ebcs=Conditions([ebc1, ebc2, ebc3]))
pb.update_materials()
return pb
def solve_problem(mesh_filename, options, comm):
order_u = options.order_u
order_p = options.order_p
rank, size = comm.Get_rank(), comm.Get_size()
output('rank', rank, 'of', size)
stats = Struct()
timer = Timer('solve_timer')
timer.start()
mesh = Mesh.from_file(mesh_filename)
stats.t_read_mesh = timer.stop()
timer.start()
if rank == 0:
cell_tasks = pl.partition_mesh(mesh, size, use_metis=options.metis,
verbose=True)
else:
cell_tasks = None
stats.t_partition_mesh = timer.stop()
output('creating global domain and fields...')
timer.start()
domain = FEDomain('domain', mesh)
omega = domain.create_region('Omega', 'all')
field1 = Field.from_args('fu', nm.float64, mesh.dim, omega,
approx_order=order_u)
field2 = Field.from_args('fp', nm.float64, 1, omega,
approx_order=order_p)
fields = [field1, field2]
stats.t_create_global_fields = timer.stop()
output('...done in', timer.dt)
output('distributing fields...')
timer.start()
distribute = pl.distribute_fields_dofs
lfds, gfds = distribute(fields, cell_tasks,
is_overlap=True,
use_expand_dofs=True,
save_inter_regions=options.save_inter_regions,
output_dir=options.output_dir,
comm=comm, verbose=True)
stats.t_distribute_fields_dofs = timer.stop()
output('...done in', timer.dt)
output('creating local problem...')
timer.start()
cells = lfds[0].cells
omega_gi = Region.from_cells(cells, domain)
omega_gi.finalize()
omega_gi.update_shape()
pb = create_local_problem(omega_gi, [order_u, order_p])
variables = pb.get_initial_state()
variables.fill_state(0.0)
variables.apply_ebc()
stats.t_create_local_problem = timer.stop()
output('...done in', timer.dt)
output('allocating global system...')
timer.start()
sizes, drange, pdofs = pl.setup_composite_dofs(lfds, fields, variables,
verbose=True)
pmtx, psol, prhs = pl.create_petsc_system(pb.mtx_a, sizes, pdofs, drange,
is_overlap=True, comm=comm,
verbose=True)
stats.t_allocate_global_system = timer.stop()
output('...done in', timer.dt)
output('creating solver...')
timer.start()
conf = Struct(method='bcgsl', precond='jacobi', sub_precond='none',
i_max=10000, eps_a=1e-50, eps_r=1e-6, eps_d=1e4,
verbose=True)
status = {}
ls = PETScKrylovSolver(conf, comm=comm, mtx=pmtx, status=status)
field_ranges = {}
for ii, variable in enumerate(variables.iter_state(ordered=True)):
field_ranges[variable.name] = lfds[ii].petsc_dofs_range
ls.set_field_split(field_ranges, comm=comm)
ev = PETScParallelEvaluator(pb, pdofs, drange, True,
psol, comm, verbose=True)
nls_status = {}
conf = Struct(method='newtonls',
i_max=5, eps_a=0, eps_r=1e-5, eps_s=0.0,
verbose=True)
nls = PETScNonlinearSolver(conf, pmtx=pmtx, prhs=prhs, comm=comm,
fun=ev.eval_residual,
fun_grad=ev.eval_tangent_matrix,
lin_solver=ls, status=nls_status)
stats.t_create_solver = timer.stop()
output('...done in', timer.dt)
output('solving...')
timer.start()
variables.apply_ebc()
ev.psol_i[...] = variables()
ev.gather(psol, ev.psol_i)
psol = nls(psol)
ev.scatter(ev.psol_i, psol)
sol0_i = ev.psol_i[...]
stats.t_solve = timer.stop()
output('...done in', timer.dt)
output('saving solution...')
timer.start()
variables.set_state(sol0_i)
out = variables.create_output()
filename = os.path.join(options.output_dir, 'sol_%02d.h5' % comm.rank)
pb.domain.mesh.write(filename, io='auto', out=out)
gather_to_zero = pl.create_gather_to_zero(psol)
psol_full = gather_to_zero(psol)
if comm.rank == 0:
sol = psol_full[...].copy()
u = FieldVariable('u', 'parameter', field1,
primary_var_name='(set-to-None)')
remap = gfds[0].id_map
ug = sol[remap]
p = FieldVariable('p', 'parameter', field2,
primary_var_name='(set-to-None)')
remap = gfds[1].id_map
pg = sol[remap]
if (((order_u == 1) and (order_p == 1))
or (options.linearization == 'strip')):
out = u.create_output(ug)
out.update(p.create_output(pg))
filename = os.path.join(options.output_dir, 'sol.h5')
mesh.write(filename, io='auto', out=out)
else:
out = u.create_output(ug, linearization=Struct(kind='adaptive',
min_level=0,
max_level=order_u,
eps=1e-3))
filename = os.path.join(options.output_dir, 'sol_u.h5')
out['u'].mesh.write(filename, io='auto', out=out)
out = p.create_output(pg, linearization=Struct(kind='adaptive',
min_level=0,
max_level=order_p,
eps=1e-3))
filename = os.path.join(options.output_dir, 'sol_p.h5')
out['p'].mesh.write(filename, io='auto', out=out)
stats.t_save_solution = timer.stop()
output('...done in', timer.dt)
stats.t_total = timer.total
stats.n_dof = sizes[1]
stats.n_dof_local = sizes[0]
stats.n_cell = omega.shape.n_cell
stats.n_cell_local = omega_gi.shape.n_cell
return stats
helps = {
'output_dir' :
'output directory',
'dims' :
'dimensions of the block [default: %(default)s]',
'shape' :
'shape (counts of nodes in x, y, z) of the block [default: %(default)s]',
'centre' :
'centre of the block [default: %(default)s]',
'2d' :
'generate a 2D rectangle, the third components of the above'
' options are ignored',
'u-order' :
'displacement field approximation order',
'p-order' :
'pressure field approximation order',
'linearization' :
'linearization used for storing the results with approximation order > 1'
' [default: %(default)s]',
'metis' :
'use metis for domain partitioning',
'save_inter_regions' :
'save inter-task regions for debugging partitioning problems',
'stats_filename' :
'name of the stats file for storing elapsed time statistics',
'new_stats' :
'create a new stats file with a header line (overwrites existing!)',
'silent' : 'do not print messages to screen',
'clear' :
'clear old solution files from output directory'
' (DANGEROUS - use with care!)',
}
def main():
parser = ArgumentParser(description=__doc__.rstrip(),
formatter_class=RawDescriptionHelpFormatter)
parser.add_argument('output_dir', help=helps['output_dir'])
parser.add_argument('--dims', metavar='dims',
action='store', dest='dims',
default='1.0,1.0,1.0', help=helps['dims'])
parser.add_argument('--shape', metavar='shape',
action='store', dest='shape',
default='11,11,11', help=helps['shape'])
parser.add_argument('--centre', metavar='centre',
action='store', dest='centre',
default='0.0,0.0,0.0', help=helps['centre'])
parser.add_argument('-2', '--2d',
action='store_true', dest='is_2d',
default=False, help=helps['2d'])
parser.add_argument('--u-order', metavar='int', type=int,
action='store', dest='order_u',
default=1, help=helps['u-order'])
parser.add_argument('--p-order', metavar='int', type=int,
action='store', dest='order_p',
default=1, help=helps['p-order'])
parser.add_argument('--linearization', choices=['strip', 'adaptive'],
action='store', dest='linearization',
default='strip', help=helps['linearization'])
parser.add_argument('--metis',
action='store_true', dest='metis',
default=False, help=helps['metis'])
parser.add_argument('--save-inter-regions',
action='store_true', dest='save_inter_regions',
default=False, help=helps['save_inter_regions'])
parser.add_argument('--stats', metavar='filename',
action='store', dest='stats_filename',
default=None, help=helps['stats_filename'])
parser.add_argument('--new-stats',
action='store_true', dest='new_stats',
default=False, help=helps['new_stats'])
parser.add_argument('--silent',
action='store_true', dest='silent',
default=False, help=helps['silent'])
parser.add_argument('--clear',
action='store_true', dest='clear',
default=False, help=helps['clear'])
options, petsc_opts = parser.parse_known_args()
comm = pl.PETSc.COMM_WORLD
output_dir = options.output_dir
filename = os.path.join(output_dir, 'output_log_%02d.txt' % comm.rank)
if comm.rank == 0:
ensure_path(filename)
comm.barrier()
output.prefix = 'sfepy_%02d:' % comm.rank
output.set_output(filename=filename, combined=options.silent == False)
output('petsc options:', petsc_opts)
mesh_filename = os.path.join(options.output_dir, 'para.h5')
dim = 2 if options.is_2d else 3
dims = nm.array(eval(options.dims), dtype=nm.float64)[:dim]
shape = nm.array(eval(options.shape), dtype=nm.int32)[:dim]
centre = nm.array(eval(options.centre), dtype=nm.float64)[:dim]
output('dimensions:', dims)
output('shape: ', shape)
output('centre: ', centre)
if comm.rank == 0:
from sfepy.mesh.mesh_generators import gen_block_mesh
if options.clear:
remove_files_patterns(output_dir,
['*.h5', '*.mesh', '*.txt'],
ignores=['output_log_%02d.txt' % ii
for ii in range(comm.size)],
verbose=True)
save_options(os.path.join(output_dir, 'options.txt'),
[('options', vars(options))])
mesh = gen_block_mesh(dims, shape, centre, name='block-fem',
verbose=True)
mesh.write(mesh_filename, io='auto')
comm.barrier()
output('field u order:', options.order_u)
output('field p order:', options.order_p)
stats = solve_problem(mesh_filename, options, comm)
output(stats)
if options.stats_filename:
from sfepy.examples.diffusion.poisson_parallel_interactive import save_stats
if comm.rank == 0:
ensure_path(options.stats_filename)
comm.barrier()
pars = Struct(dim=dim, shape=shape, order=options.order_u)
pl.call_in_rank_order(
lambda rank, comm:
save_stats(options.stats_filename, pars, stats, options.new_stats,
rank, comm),
comm
)
if __name__ == '__main__':
main()